

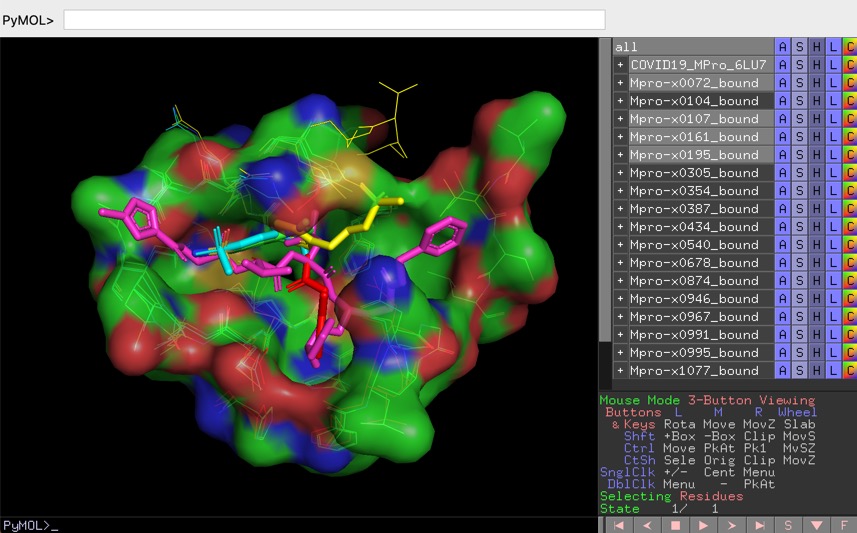
To begin with, I will reproduce the figure that appears on the September 2000 entry of MotM, corresponding to hen egg white lysozyme (PDB ID: 2LYZ).įirst load the structure of 2LYZ (for example by using fetch 2lyz),
#Pymol tutorial español how to#
At first I just wanted to get an idea of how to do some figures with Goodsell’s style, but it became kind of an obsession. However, I took some ideas from them and managed to develop two methods using PyMOL. There are several tutorials and tools to emulate his rendering style 1, but none of these fully convince me. To generate the figures in MoTM Goodsell uses some software he developed (apparently in Fortran, according to this German link). However, reproducing the way he renders molecules is not a straightforward task. Like many people, I like the unique style in which David Goodsell illustrates the structures of the Molecule of the Month (MotM) section in the Protein Data Bank. In particular, I am fascinated by the various ways of representing three-dimensional structures of biological macromolecules, either by computer graphics, animations, or tangible, physical models. cmd file, and modify it.Versión en español: Aplicando ingeniería reversa a David Goodsell DecemReverse engineering David Goodsell Introductionįor some time I have been interested in the relationship between art and structural biology. mkdir %HOMEPATH%\pymol Then make sure, PyMOL starts here, when you open the shortcut. Now start pymol, and enjoy all the plugins available from the menu. Then "File->Save as->All files-> C:\Python27\Lib\site-packages\pymol\pymol_path\run_on_startup.py

environ ) # Make setting changes to Plugin Manager import ugins pymol. # Add paths to sys.path so PyMOL can find modules and scripts import sys, os pymol_git = os. Navigate to the installation directory in a CMD window
Open-Source PyMOL is available free of charge.
#Pymol tutorial español movie#
The bundle also includes ready-to-use APBS, RigiMOL, an MPEG encoder for movie export, and a small molecule energy minimization engine.
#Pymol tutorial español .exe#
Schrödinger provides an installer to paying sponsors (EXE for PyMOL 2.0, MSI for previous version).


 0 kommentar(er)
0 kommentar(er)
